Single Template PCR Reaction This technique uses a single template which can be a genomic DNA along with several pairs of forward and reverse primers to amplify specific regions within a template. When a primer annotation is added to a sequence the primer_bind portion of the primer will be colored Dark Green if it binds in the forward direction ie.

Design Of Forward And Reverse Primers The Synthesized Primers Are Download Scientific Diagram

Forward And Reverse Sense And Antisense Primers Youtube

How To Select Primers For Polymerase Chain Reaction
MethPrimer - An online program for designing PCR primers for mapping DNA methylation such as MSP and BSP PCR and for predicting CpG islands.
How to design forward and reverse primers for pcr. Use this online T m calculator with values of 50 mM for salt concentration and 300 nM for oligonucleotide concentration. DNA polymerase starts synthesizing new DNA from the 3 end of the primer. GAC CCC AAA ATC AGC GAA AT.
Your chosen restriction site for cloning usually 6-8bp Hybridization Sequence. Name Description Oligonucleotide Sequence 53 Label 1 Final Conc. PCR polymerase chain reaction is an invaluable tool for molecular biology research.
In Geneious Prime various operations can add primers to target sequences as annotations for example using the tool Primers Test with Saved Primers covered in detail in Exercise 6. Design your PCR primers to conform to the following guidelines. The polymerase chain reaction PCR uses a pair of custom primers to direct DNA elongation toward each other at opposite ends of the sequence being amplified.
When designing primers follow these guidelines. Also if you are performing a one-step reverse transcription PCR RT. Reverse transcription polymerase chain reaction RT-PCR is a laboratory technique combining reverse transcription of RNA into DNA in this context called complementary DNA or cDNA and amplification of specific DNA targets using polymerase chain reaction PCR.
2019-Novel Coronavirus 2019-nCoV Real-time rRT-PCR Panel Primers and Probes. Primers are synthetic DNA strands of about 18 to 25 nucleotides complementary to 3 end of the template strand. PrimerX is a web-based program written to automate the design of mutagenic PCR primers for site-directed mutagenesis.
TCT GGT TAC TGC CAG TTG AAT CTG. Adjust primer locations so they are. Add the forward and reverse primers to the thinned walled PCR tubes.
The process of designing specific primers typically involves two stages. For real-time PCR we want to use an annealing temperature of around 60 o C for our PCR reaction. CDNA 2 x SYBR Green mix 2 qRT-PCR conditions and instrument setup a.
Primer-BLAST allows users to design new target-specific primers in one step as well as to check the specificity of pre-existing primers. Primer-BLAST also supports placing primers based on exonintron locations and excluding single nucleotide polymorphism SNP sites in primers. One way to calculate T m values is by using the nearest-neighbor method.
Designing primers for PCR based cloning. Open the Mx4000 program and set up. The forward and reverse primer populations.
QPCR primers that anneal poorly or that anneal to more than one sequence during amplification can significantly impact the quality and reliability of your results. When regular primers are synthesised only two populations of primers are present in the PCR reaction. Design primers that have a GC content of 5060.
Primers can be designed as regular or degenerate. Strive for a T m between 50 and 65C. The incorporation of LNA in both the forward and reverse PCR amplification primers makes it possible to design assays that can distinguish between miRNA sequences that differ by only one nucleotide see figure Single-nucleotide discrimination.
The region of the primer that binds to the sequence to be amplified usually 18-21bp. However the most important considerations for primer design should be their T m value and specificity. Forward and reverse primers are the two types of primers used in the PCR polymerase chain reaction to amplify a specific part of a DNA strand.
Select dual-labeled probes and unlabeled sequence detection primers for real-time PCR applications using TaqMan probe-based chemistry. The forward primer and the reverse primer. The PrimerQuest Tool makes it easy to design basic and highly customized primers and probes for PCR and qPCR.
Two primers must be designed for PCR. Use primer final concentration of 200nM. It is used in laboratories around the world in a wide array of applications such as cloning gene expression analysis genotyping sequencing and mutagenesis.
To design primers manually highlight a region of template DNA on the sequence map by selecting the desired range right clicking and choosing to create either the forward or reverse primer. Do qRT-PCR and test the selected primers 1 qRT-PCR set up. Multiple Template PCR Reaction It uses multiple templates and several primer sets in.
Primers should also be free of strong secondary structures and self-complementarity. The basic PCR primers for molecular cloning consist of. DNA-based Protein-based Primer Characterization Documentation Links.
Specific amplification of the intended target requires that primers do not have matches to other targets in certain orientations and within certain distances that allow undesired amplification. IDT recommends that you aim for PCR primers between 18 and 30 bases. Key Areas Covered 1.
Do two reactions for each pair of primers by using cDNA and H2O as templates separately. Furthermore 5 primers refer to forward primers while 3 primers refer to reverse primers. Degenerate primer design requires a set of reference sequences.
Do this before adding the master mix so you know that the primers have been added pipet the forward primer onto one side of the tube wall and the reverse primer onto the other. Now that you understand the fundamentals of using this design tool look for future articles from IDT about experiments requiring more complex assay design considerations such as splice-specific multiplexing SNPs and CNV designs. It complements the reverse strand or light green.
Several software programs such as In-Silico PCR and Reverse ePCR do not design primers but rather determine the amplification targets of user-supplied primer pairs. Choosing appropriate primers is probably the single most important factor affecting the polymerase chain reaction PCR. The Max Tm difference is the difference in Tm between the forward and reverse primers which is currently set as 3 o C.
Based on your input PrimerX compares a template DNA sequence with a DNA or protein sequence that already. In addition the assays can discriminate between mature miRNA sequences and precursor miRNA. PCR primer design.
However even with the help of these tools finding specific primers can still be a difficult process because users often need to go through many candidate primers manually. Add the master mix to the thin walled PCR tubes. Quantitative PCR qPCR primer design is a critical step when setting up your qPCR or reverse transcription-qPCR assay RT-qPCR for gene expression analysis.
Extra base pairs on the 5 end of the primer assist with restriction enzyme digestion usually 3-6bp Restriction Site. Phosphoramidites for Nucleic Acid Synthesis Synthesize high-quality DNA and RNA oligos with the confidence from partnering with a. These primers are typically between 18 and 24 bases in length and must code for only the specific upstream and downstream sites of the sequence being amplified.
What are Forward Primers Definition Features Importance 2. All procedures should be done on ice. This is achieved by monitoring the amplification reaction using.
When degenerate primers are synthesized there may be multiple forward and reverse. It is primarily used to measure the amount of a specific RNA.

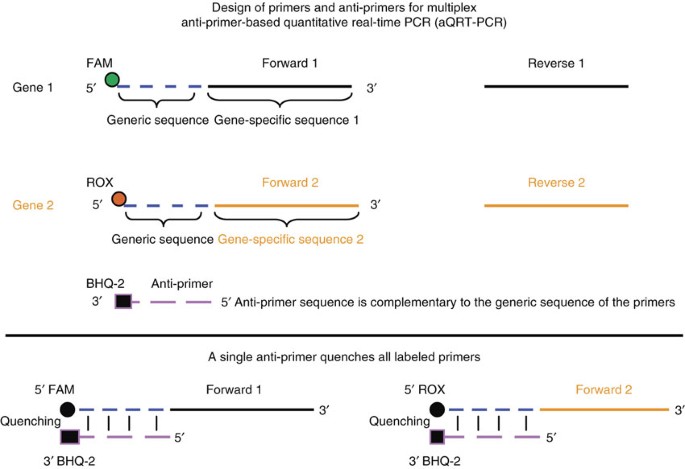
Anti Primer Quenching Based Real Time Pcr For Simplex Or Multiplex Dna Quantification And Single Nucleotide Polymorphism Genotyping Nature Protocols

Solved 16 Design The Forward And Reverse Primers To Mutate Chegg Com
Overhang Pcr
How To Make Primers For Pcr Pediaa Com
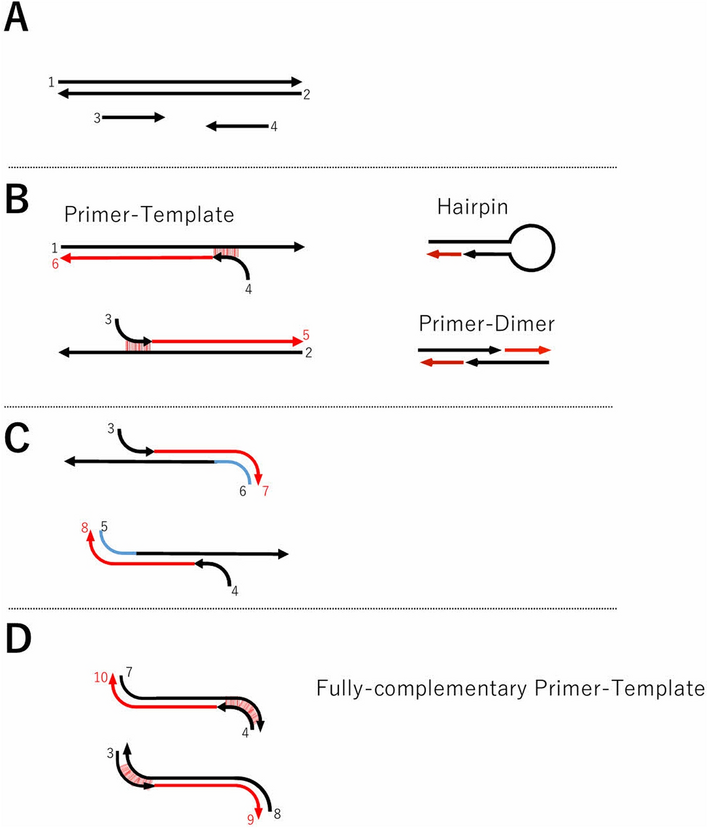
Prediction Of Pcr Amplification From Primer And Template Sequences Using Recurrent Neural Network Scientific Reports

Primer Design Synthesis Dna And Cloning Services Research Cro Custom Services
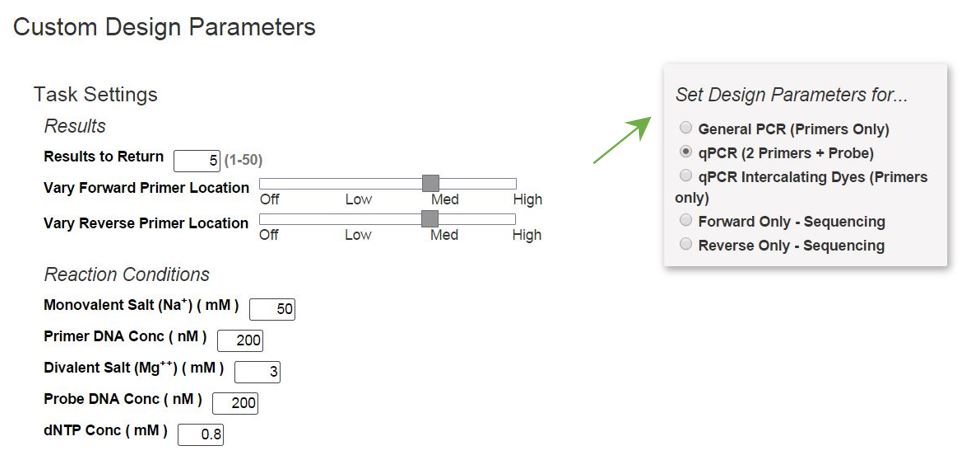
Primer Design Tools For Pcr Qpcr Idt

Barcoded Library Preparation Strategy Forward And Reverse Pcr Primers Download Scientific Diagram
